
Features
Production
Research
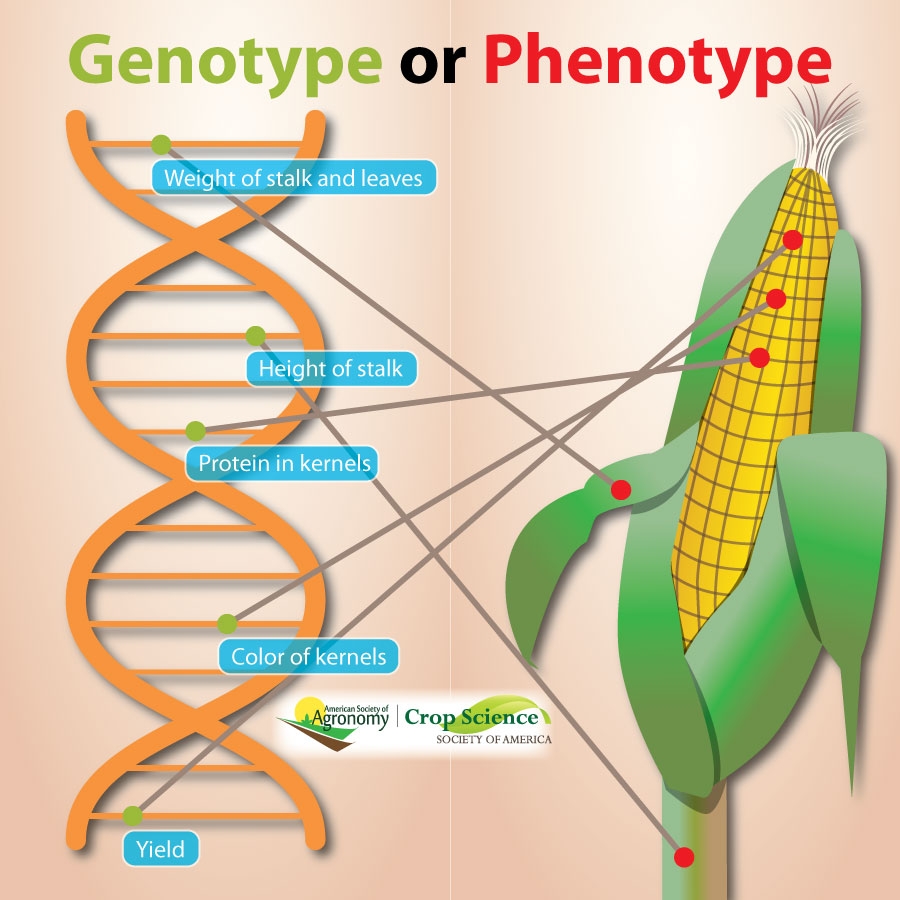
How do breeders know which part of the DNA corresponds to the trait they are breeding for?
December 31, 2018 By American Society of Agronomy and Crop Science Society of America
 ASA/CSSA
ASA/CSSAPlant breeders need to know there’s good genetics in the crops they are developing. The recent Sustainable, Secure Food blog post explains how crop scientists improve crops using data gathered from both the field and the lab.
“DNA is like an ‘instruction manual’ that gives every living plant and animal its own unique traits,” says Christine Bradish, AVOCA.
For plants, this can include traits that can be measured in the field—for example, flower color, plant height, or disease resistance. These are phenotypes. Researchers take as many measurements as possible, and across the widest range of field locations and years as possible.
On a microscopic level in the lab, scientists can map out a plant’s traits in the chromosomes. This is the genotype. “Many plants now have entire genetic maps done, or better yet, have all their DNA sequenced. It’s a big deal to have a plant’s DNA sequenced, and can help breeders in selecting new crop varieties to develop.”
Both phenotypes and genotypes are important to plant breeders, “Crop breeders often are working to find plants with new traits. This could be resistance to one particular pest, like rust resistance. Or, we can be looking for ways to increase yield, which is a very important trait. So, we are breeding for improved phenotypes–and we can look at genotypes to predict outcomes of our breeding work.” | For the full story, CLICK HERE.
Print this page